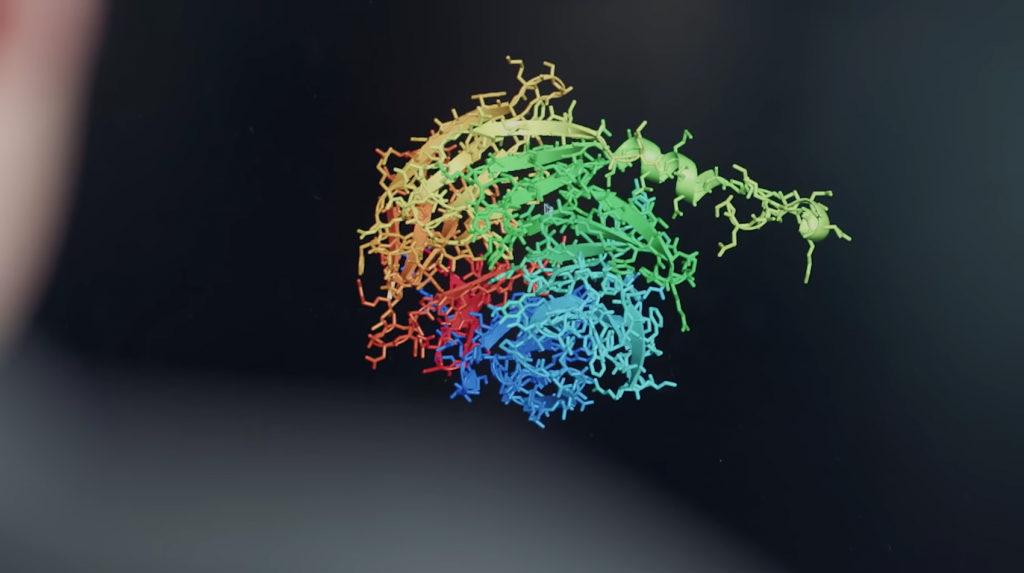
The AlphaFold team runs a simulation of protein folding in a video released by DeepMind. (Source: YouTube)
DeepMind Pinpoints Protein Folding Structures, Solving One of the Greatest Problems in Medical Research
International scientists are celebrating the recent announcement by DeepMind of its breakthrough in protein folding using its AlphaFold AI system. A story on USA Today explains how the 50-year research project was supercharged by the AI program and input from teams of hundreds of researchers. The advances could accelerate the development of everything from medical drugs and vaccines to understanding and curing diseases.
“This computational work represents a stunning advance on the protein-folding problem, a 50-year old grand challenge in biology,” said Venki Ramakrishnan, president of the U.K.’s Royal Society. “It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research.”
The London-based AI firm, which was founded in 2010 and developed AlphaGo, joined forces with Google in 2014. It has developed other advances in neuroscience, machine learning, robotics, language, computer vision and environments—but none have the potential to impact humankind more than understanding protein folding. Alzheimer’s and Parkinson’s diseases are among many which could be solved by this technology.
“Many of the greatest challenges facing society, like developing treatments for diseases or finding enzymes that break down industrial waste, are fundamentally tied to proteins and the role they play,” DeepMind told USA TODAY.
According to a story on Nature.com, AlphaFold far outperformed lab experiments in predicting protein structures. Scientists spoke exuberantly about the news, which was confirmed when the AI team outperformed around 100 other teams in a biennial protein-structure prediction challenge called CASP, short for Critical Assessment of Structure Prediction. CASP held its conference virtually on November 30.
“This is a big deal,” John Moult told Nature. He is a computational biologist at the University of Maryland in College Park who co-founded CASP in 1994 to improve computational methods for accurately predicting protein structures. “In some sense the problem is solved.”
“’It’s a game changer,’ Andrei Lupas, an evolutionary biologist at the Max Planck Institute for Developmental Biology in Tübingen, Germany, told Nature. He assessed the performance of different teams in CASP. ‘This will change medicine. It will change research. It will change bioengineering. It will change everything.’ AlphaFold has already helped him find the structure of a protein that has vexed his lab for a decade, and he expects it will alter how he works and the questions he tackles.”
A story on MIT’s technologyreview.com confirms that AlphaFold has far exceeded expectations, quoting Mohammed AlQuraishi, a systems biologist at Columbia University who has developed his own software for predicting protein structure, as thinking it would take researchers 10 years to get from AlphaFold’s 2018 results to those it achieved in 2020.
“This is close to the physical limit for how accurate you can get, AlQuraishi said. “These structures are fundamentally floppy. It doesn’t make sense to talk about resolutions much below that.”
AlQuraishi’s system, which uses an algorithm called a recurrent geometrical network (RGN), can find protein structures a million times faster—returning results in seconds rather than days, but its predictions are less accurate.
read more at deepmind.com





Leave A Comment